Factor Variable and Random Effects Plots
plotblock.RdFunction to plot effects for model terms including factor, or group variables for random effects.
plotblock(x, residuals = FALSE, range = c(0.3, 0.3),
col.residuals = "black", col.lines = "black", c.select = NULL,
fill.select = NULL , col.polygons = NULL, data = NULL,
shift = NULL, trans = NULL, labels = NULL, ...)Arguments
- x
Either a
listof length of the unique factors, where eachlistelement contains the estimated effects for one factor as a matrix, or one data matrix with first column as the group or factor variable. Also formulas are accepted, e.g it is possible to specify the plot withf ~ xorf1 + f2 ~ x. By convention, the covariate for which effects should be plotted, is always in the first column in the resulting data matrix, that is used for plotting, i.e. in the second formula example, the data matrix iscbind(x, f1, f2), also see argumentc.selectandfill.select.- residuals
If set to
TRUE, residuals will be plotted if available. Residuals may be set as anattribute ofxnamed"residuals", where the residuals must be a matrix with first column specifying the covariate, and second column the residuals that should be plotted.- range
Numeric vector, specifying the left and right bound of the block.
- col.residuals
The color of the partial residuals.
- col.lines
Vector of maximum length of columns of
xminus 1, specifying the color of the lines.- c.select
Integer vector of maximum length of columns of
x, selects the columns of the resulting data matrix that should be used for plotting. E.g. ifxhas 5 columns, thenc.select = c(1, 2, 5)will select column 1, 2 and 5 for plotting. Note that first element ofc.selectshould always be 1, since this is the column of the covariate the effect is plotted for.- fill.select
Integer vector, select pairwise the columns of the resulting data matrix that should form one polygon with a certain background color specified in argument
col. E.g.xhas three columns, or is specified with formulaf1 + f2 ~ x, then settingfill.select = c(0, 1, 1)will draw a polygon withf1andf2as boundaries. Ifxhas five columns or the formula is e.g.f1 + f2 + f3 + f4 ~ x, then settingfill.select = c(0, 1, 1, 2, 2), the pairsf1,f2andf3,f4are selected to form two polygons.- col.polygons
Specify the background color for the upper and lower confidence bands, e.g.
col = c("green", "red").- data
If
xis a formula, adata.frameorlist. By default the variables are taken fromenvironment(x): typically the environment from whichplotblockis called.- shift
Numeric constant to be added to the smooth before plotting.
- trans
Function to be applied to the smooth before plotting, e.g., to transform the plot to the response scale.
- labels
Character, labels for the factor levels.
- ...
Graphical parameters, please see the details.
Details
Function plotblock draws for every factor or group the effect as a "block" in one graphic,
i.e., similar to boxplots, estimated fitted effects, e.g., containing quantiles of MCMC samples,
are drawn as one block, where the upper lines represent upper quantiles, the
middle line the mean or median, and lower lines lower quantiles, also see the examples. The
following graphical parameters may be supplied additionally:
cex: Specify the size of partial residuals,lty: The line type for each column that is plotted, e.g.lty = c(1, 2),lwd: The line width for each column that is plotted, e.g.lwd = c(1, 2),poly.lty: The line type to be used for the polygons,poly.lwd: The line width to be used for the polygons,densityangle,border: Seepolygon,...: Other graphical parameters, see functionplot.
Examples
## Generate some data.
set.seed(111)
n <- 500
## Regressors.
d <- data.frame(fac = factor(rep(1:10, n/10)))
## Response.
d$y <- with(d, 1.5 + rnorm(10, sd = 0.6)[fac] +
rnorm(n, sd = 0.6))
if (FALSE) ## Estimate model.
b <- bamlss(y ~ s(fac,bs="re"), data = d)
summary(b)
#> Error in eval(expr, envir, enclos): object 'b' not found
## Plot random effects.
plot(b)
#> Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'plot': object 'b' not found
## Extract fitted values.
f <- fitted(b, model = "mu", term = "fac")
#> Error in eval(expr, envir, enclos): object 'b' not found
f <- cbind(d["fac"], f)
#> Error in eval(expr, envir, enclos): object 'f' not found
## Now use plotblock.
plotblock(f)
#> Error in eval(expr, envir, enclos): object 'f' not found
## Variations.
plotblock(f, fill.select = c(0, 1, 0, 1), col.poly = "red")
#> Error in eval(expr, envir, enclos): object 'f' not found
plotblock(f, fill.select = c(0, 1, 0, 1), col.poly = "lightgray",
lty = c(2, 1, 2), lwd = c(2, 1, 2))
#> Error in eval(expr, envir, enclos): object 'f' not found
## More examples.
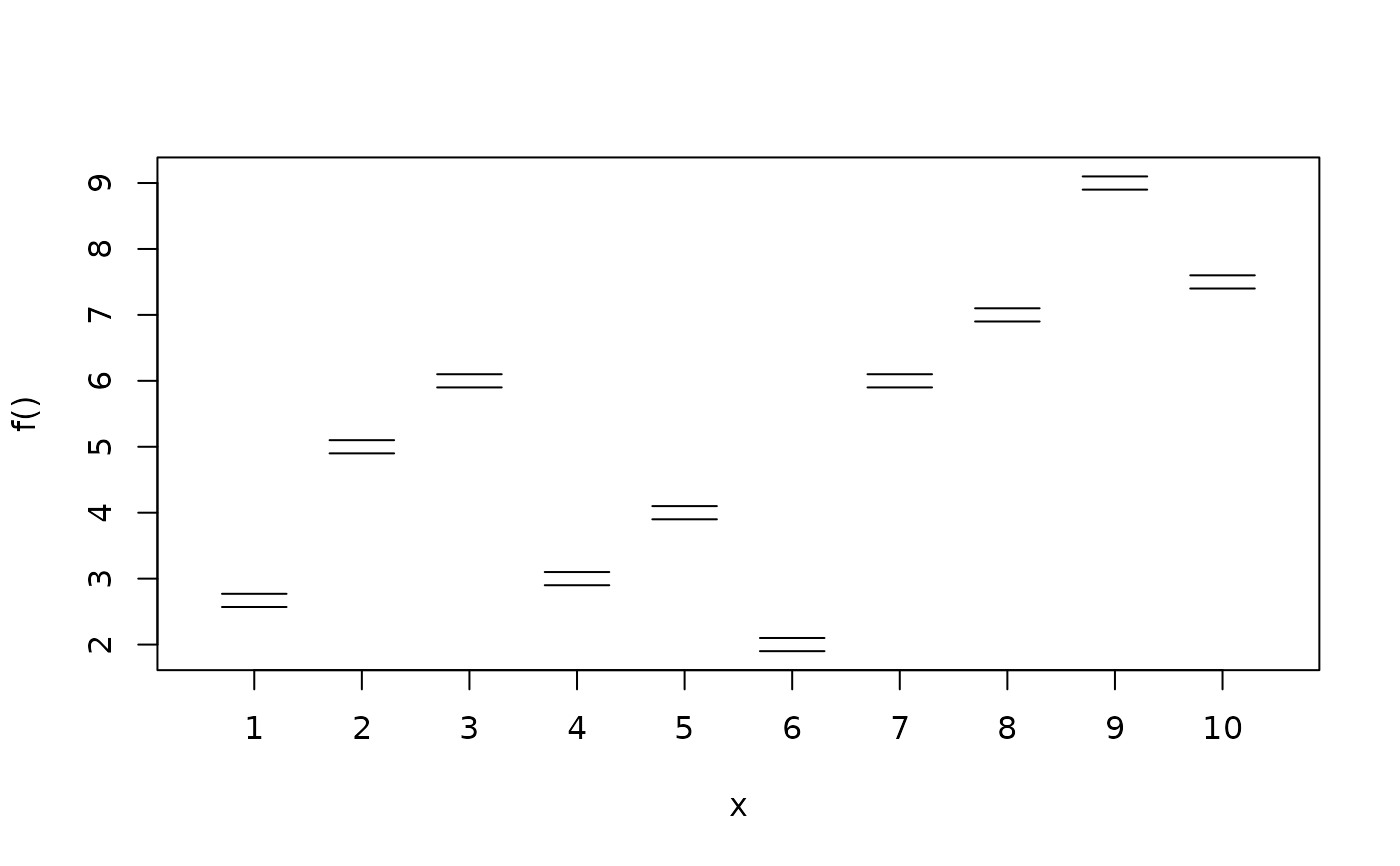
plotblock(y ~ fac, data = d, range = c(0.45, 0.45))
 d <- data.frame(fac = factor(rep(1:10, n/10)))
d$y <- with(d, c(2.67, 5, 6, 3, 4, 2, 6, 7, 9, 7.5)[fac])
plotblock(y ~ fac, data = d)
d <- data.frame(fac = factor(rep(1:10, n/10)))
d$y <- with(d, c(2.67, 5, 6, 3, 4, 2, 6, 7, 9, 7.5)[fac])
plotblock(y ~ fac, data = d)
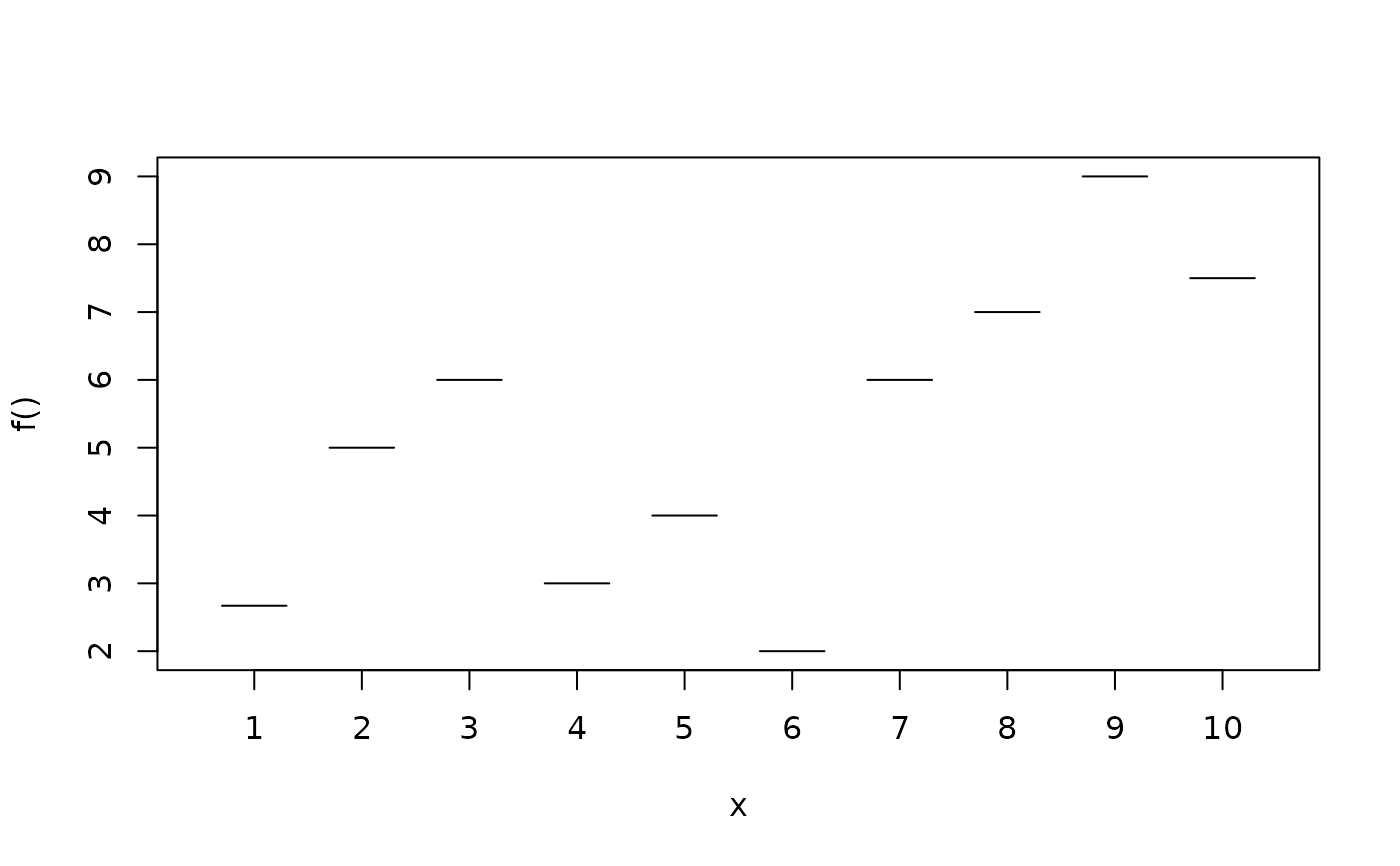 plotblock(cbind(y - 0.1, y + 0.1) ~ fac, data = d)
plotblock(cbind(y - 0.1, y + 0.1) ~ fac, data = d)